Our 3D Platform Technology
Scalable, fast, quantitative and in vivo. Analyze conformational changes across the whole proteome- for example to identify drug targets, characterize drug target engagement, identify off-targets, or to better characterize a disease state.
By capturing the native conformation of proteins in living cells in a fast, scalable, and quantitative way, CPP makes it easier than ever to identify conformational changes across the whole proteome and accelerate your research.
Background
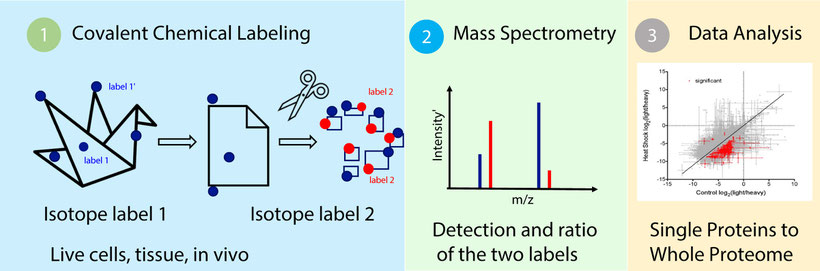
Think of a protein as an origami. The folded structure provides the function. Changes to the fold change the function and occur naturally in response to internal and external stimuli, but also upon ligand or drug binding, for example. Our core technology -Covalent Protein Painting (CPP)- allows to identify and quantify such conformational changes of proteins for the first time in live cells across the whole proteome. Based on small, cell permeable, isotope encoded covalent labels that are read out in a high-resolution mass spectrometer, identity and quantity of a conformational change can be accurately and robustly determined. In particular, solvent accessible lysines are labeled with an isotope encoded, cell permeable label in live cells, while solvent-excluded lysines, "hidden" inside the protein structure, are labeled only upon denaturing and digestion of the proteins. The first and second label differ in their isotope composition and the ratio of the two labels can be measured in a standard high resolution mass spectrometer and reflects the fold of the corresponding protein. Changes in the ratio of a site between two samples represent a conformational change of a protein and can be directly compared between samples. Thus, the 3D proteome platform can be applied to analyze drug target engagement, to identify off-targets and toxic effects in toxicology assays and for structure-function analysis.
Efficient. Greater than 98% labeling efficiency is achieved with the amine reactive labels (ref 1).
Fast. Complete labeling can be achieved in less than 20 seconds (ref 1).
Compatible. CPP labeling can be analyzed with standard high resolution mass spectrometers with a resolution > 30,000. Resulting data can be analyzed with standard proteomics software packages such as Proteome Discoverer.
References
- Bamberger C, Pankow S, Martinez-Bartolome S, Ma M, Diedrich J, Rissman RA, Yates JR, 3rd. Protein Footprinting via Covalent Protein Painting Reveals Structural Changes of the Proteome in Alzheimer's Disease. J Proteome Res. 2021;20(5):2762-71. Epub 2021/04/20. PubMed PMID: 33872013.
- Son A, Pankow S, Bamberger C, Yates JR, 3rd. Quantitative structural proteomics in living cells by covalent protein painting. Methods in Enzymology, Volume 679, 2023, Pages 33-63, ISBN 9780323992640, https://doi.org/10.1016/bs.mie.2022.08.046.
- Pankow S, Bamberger C, Martínez-Bartolomé S, Park SK, Yates JR, 3rd. Identification of in vivo CFTR conformations during biogenesis and upon misfolding by covalent protein painting (CPP). bioRxiv 2021.03.02.433670; doi:https://doi.org/10.1101/2021.03.02.433670.
- Bamberger C, Diedrich J, Martínez-Bartolomé S, Park SK, Yates JR, 3rd. Cancer Conformational Landscape Shapes Tumorigenesis. J Proteome Res. 2022;21(4):1017-1028. Epub 2022/03/10.